
s, -samples Process only the listed samples. r, -regions Process the given regions (comma-separated list or one region per line in a file). R, -refseq Compare the actual sequence, not just positions. per-chromosome outputs from vcf-compare) can be given. Use zero based indecies if field has more than one value.

The names are colon separated and mustĪppear in the same order as the files on the command line. The argument to this options is aĬomma-separated list or one mapping per line in a file. m, -name-mapping Use with -g when comparing files with differing column names. ignore-indels Exclude sites containing indels from genotype comparison g, -cmp-genotypes Compare genotypes, not only positions Giving the option multiple times increases verbosity Please do not use as it will be dropped in the future. c, -chromosomes Same as -r, left for backward compatibility. a, -apply-filters Ignore lines where FILTER column is anything else than PASS or '.' Vcf-compare About: Compare bgzipped and tabix indexed VCF files. u, -unique-messages Output all messages only once. d, -duplicates Warn about duplicate positions. $ bcftools reheader -h heads.txt |bcftools view|less $ bcftools norm -d all -Oz -o data_remove_ # snps|indels|both|all|exact $ bcftools view -v snps -Oz -o data_only_ Manipulation $ bcftools view -I -Oz -o data_remove_ $ bcftools view data.vcf -Oz -o & bcftools index -t # .tbi $ bcftools view data.vcf -Oz -o & bcftools index (-c) # default : -c, .csi
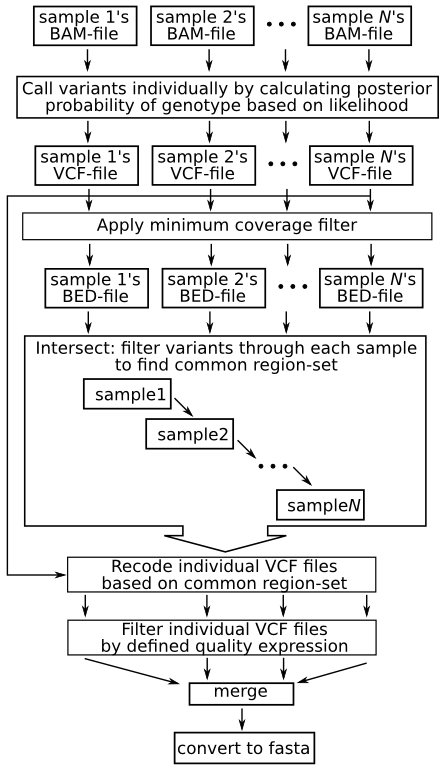
$ bgzip -c data.vcf > & tabix -p vcf # .tbi All commands work transparently with both VCFs and BCFs, both uncompressed and BGZF-compressed. BCFtools is a set of utilities that manipulate variant calls in the Variant Call Format (VCF) and its binary counterpart BCF.


 0 kommentar(er)
0 kommentar(er)
